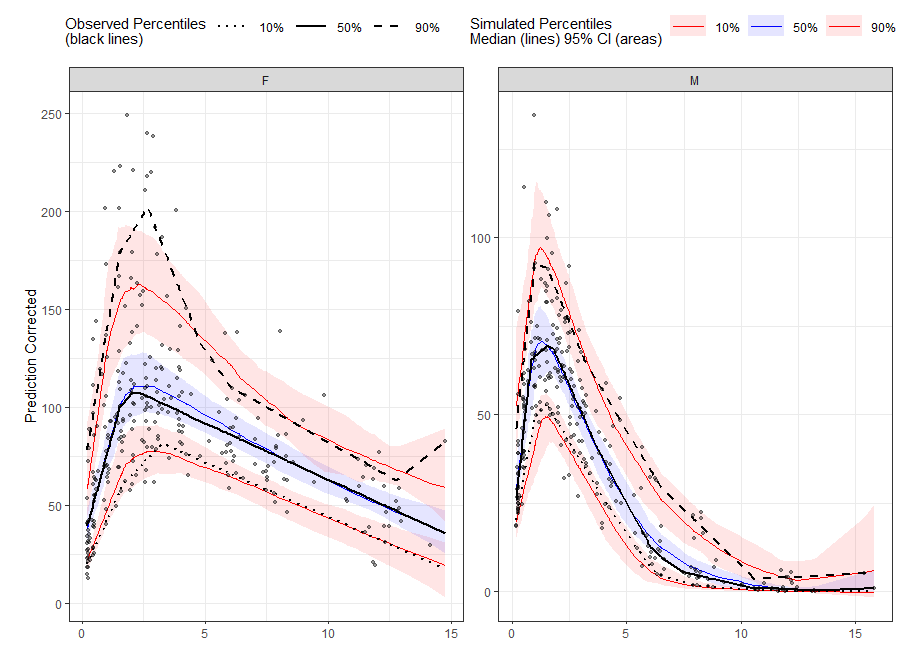
VPC Percentiles and Prediction Intervals.
tidyvpc 
Overview
tidyvpc provides a flexible and comprehensive toolkit for parameterizing a Visual Predictive Check (VPC) in R. With tidyverse style syntax, you can chain together functions (e.g., %>% or |>) to easily perform stratification, censoring, prediction correction, and more. tidyvpc supports both continuous and categorical VPC.
Installation and Running information
# CRAN
install.packages("tidyvpc")
# Development
# If there are errors (converted from warning) during installation related to packages
# built under different version of R, they can be ignored by setting the environment variable
# R_REMOTES_NO_ERRORS_FROM_WARNINGS="true" before calling remotes::install_github()
Sys.setenv(R_REMOTES_NO_ERRORS_FROM_WARNINGS="true")
remotes::install_github("certara/tidyvpc")
Learning tidyvpc
The Certara.VPCResults package offers a Shiny app that can be used to easily generate the underlying tidyvpc and ggplot2 code used to create your VPC.
After importing the observed and simulated data into your R environment, use the function vpcResultsUI() to parameterize the VPC and customize the resulting plot output using the Shiny GUI - then generate the R code to reproduce from command line!
install.packages("Certara.VPCResults",
repos = c("https://certara.jfrog.io/artifactory/certara-cran-release-public/",
"https://cloud.r-project.org"),
method = "libcurl")
library(tidyvpc)
library(Certara.VPCResults)
vpcResultsUI(observed = obs_data[MDV == 0], simulated = sim_data[MDV == 0])
The Shiny application can serve as a learning heuristic and ensures reproducibility by allowing you to save R and/or Rmd scripts. Additionally, you may render RMarkdown to an html, pdf, or docx output report. Click here to learn more about Certara.VPCResults.
Data Preprocessing
tidyvpc requires a specific structure of observed and simulated data in order to successfully generate VPC.
- DV cannot be missing in observed/simulated data i.e. subset
MDV == 0 - Observed data must be ordered by: Subject-ID, IVAR (Time)
- Simulated data must be ordered by: Replicate, Subject-ID, IVAR (Time)
See tidyvpc::obs_data and tidyvpc::sim_data for example data structures.
Usage
library(magrittr)
library(ggplot2)
library(tidyvpc)
# Filter MDV = 0
obs_data <- tidyvpc::obs_data[MDV == 0]
sim_data <- tidyvpc::sim_data[MDV == 0]
#Add LLOQ for each Study
obs_data$LLOQ <- obs_data[, ifelse(STUDY == "Study A", 50, 25)]
# Binning Method on x-variable (NTIME)
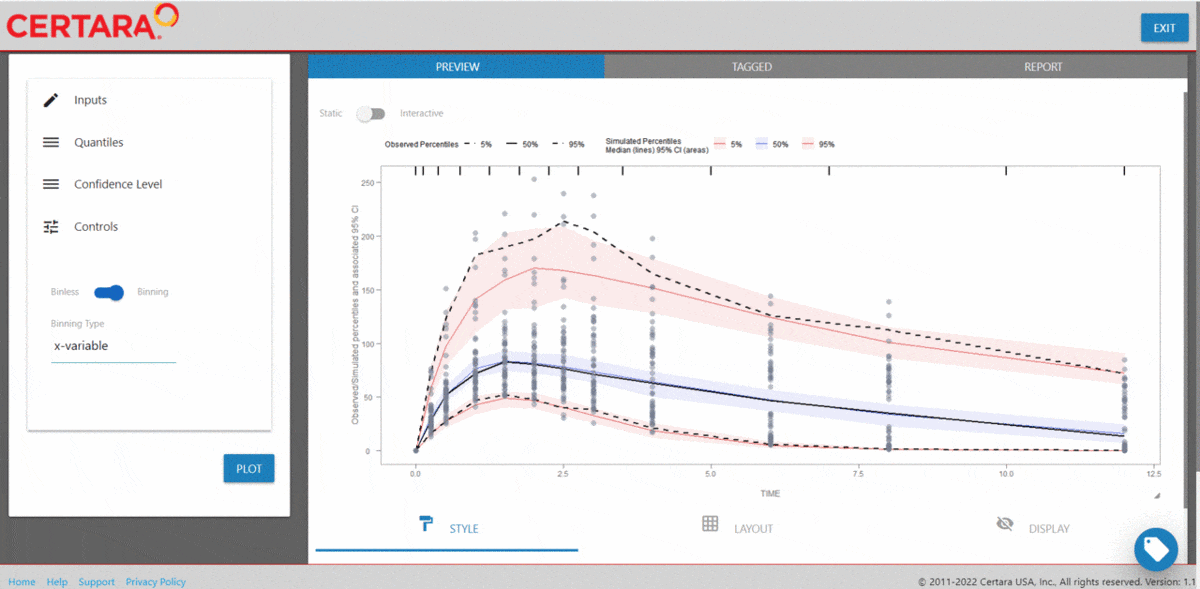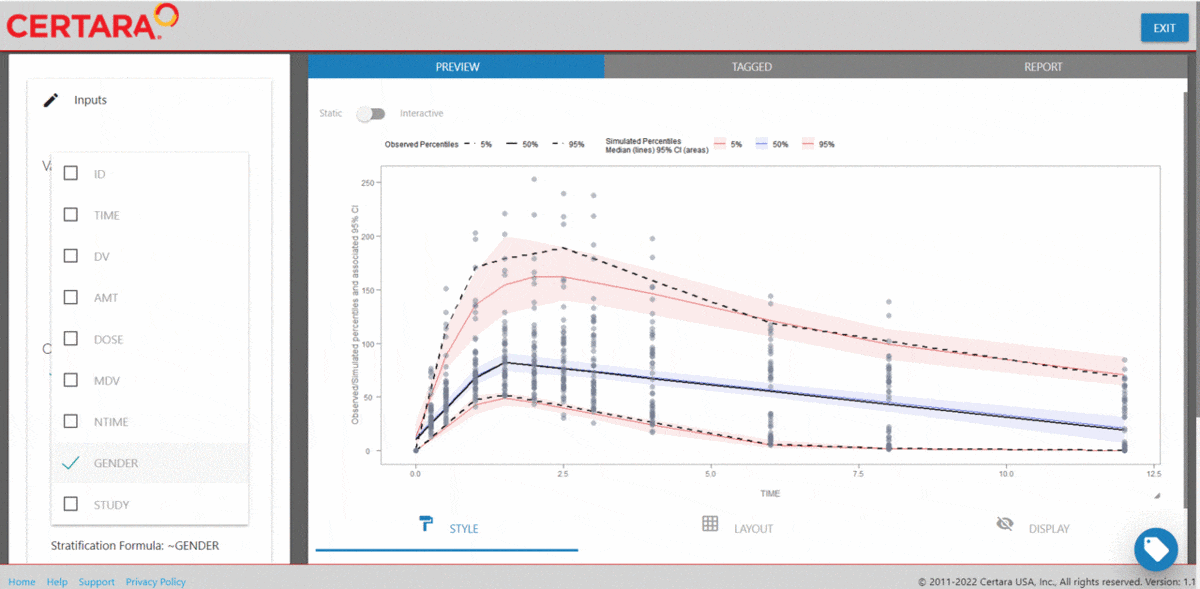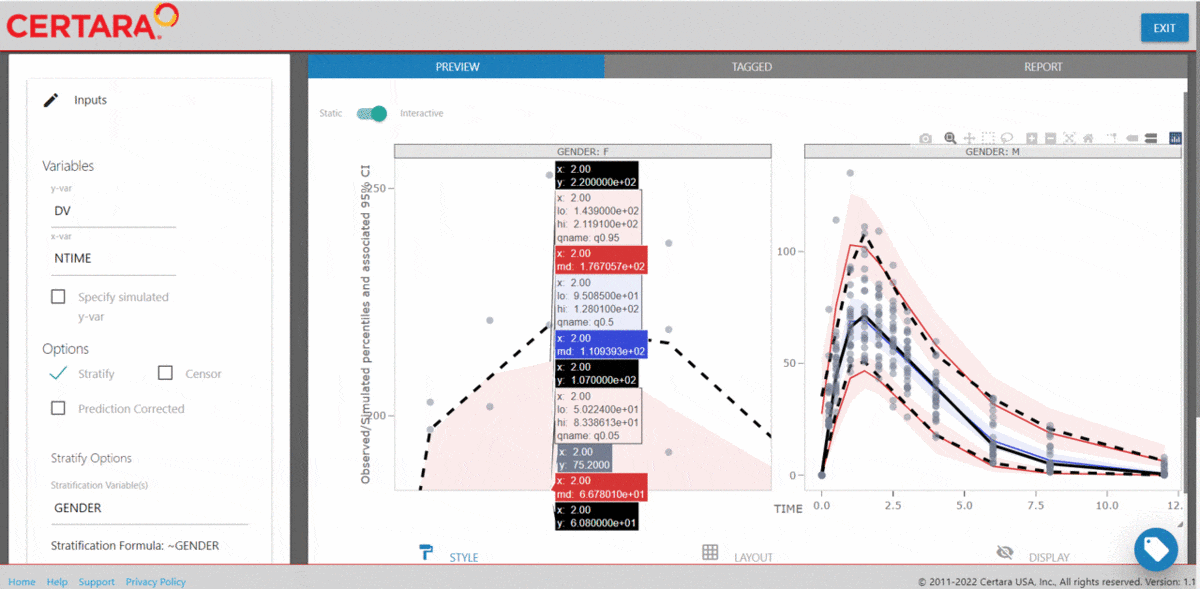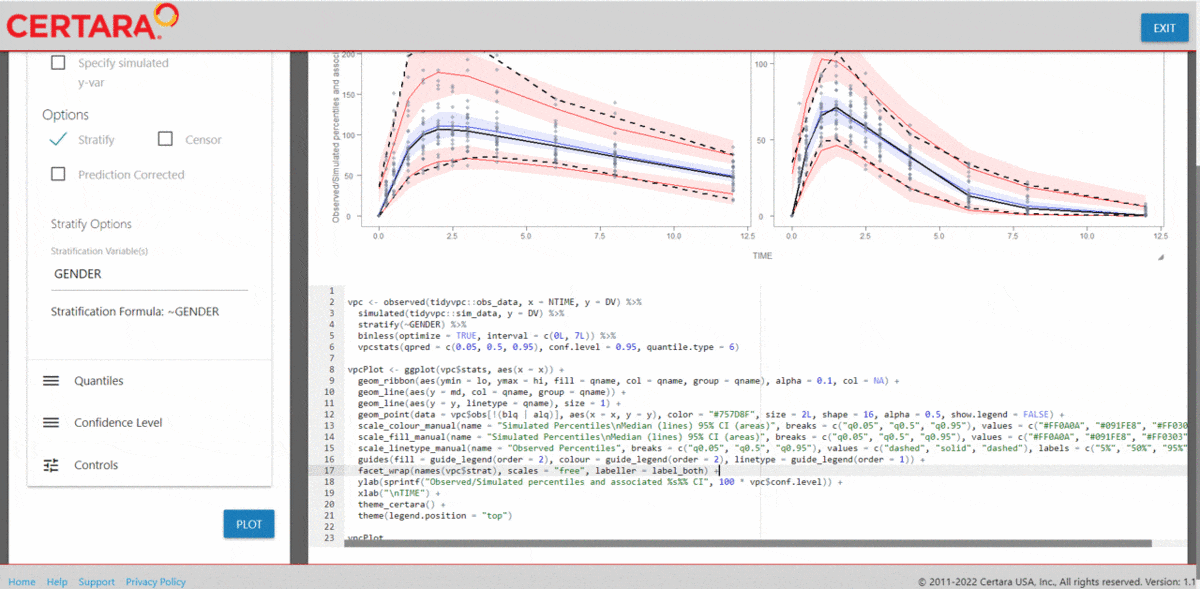
vpc <- observed(obs_data, x=TIME, y=DV) %>%
simulated(sim_data, y=DV) %>%
censoring(blq=(DV < LLOQ), lloq=LLOQ) %>%
stratify(~ STUDY) %>%
binning(bin = NTIME) %>%
vpcstats()
Plot Code:
ggplot(vpc$stats, aes(x=xbin)) +
facet_grid(~ STUDY) +
geom_ribbon(aes(ymin=lo, ymax=hi, fill=qname, col=qname, group=qname), alpha=0.1, col=NA) +
geom_line(aes(y=md, col=qname, group=qname)) +
geom_line(aes(y=y, linetype=qname), size=1) +
geom_hline(data=unique(obs_data[, .(STUDY, LLOQ)]),
aes(yintercept=LLOQ), linetype="dotted", size=1) +
geom_text(data=unique(obs_data[, .(STUDY, LLOQ)]),
aes(x=10, y=LLOQ, label=paste("LLOQ", LLOQ, sep="="),), vjust=-1) +
scale_colour_manual(
name="Simulated Percentiles\nMedian (lines) 95% CI (areas)",
breaks=c("q0.05", "q0.5", "q0.95"),
values=c("red", "blue", "red"),
labels=c("5%", "50%", "95%")) +
scale_fill_manual(
name="Simulated Percentiles\nMedian (lines) 95% CI (areas)",
breaks=c("q0.05", "q0.5", "q0.95"),
values=c("red", "blue", "red"),
labels=c("5%", "50%", "95%")) +
scale_linetype_manual(
name="Observed Percentiles\n(black lines)",
breaks=c("q0.05", "q0.5", "q0.95"),
values=c("dotted", "solid", "dashed"),
labels=c("5%", "50%", "95%")) +
guides(
fill=guide_legend(order=2),
colour=guide_legend(order=2),
linetype=guide_legend(order=1)) +
theme(
legend.position="top",
legend.key.width=grid::unit(1, "cm")) +
labs(x="Time (h)", y="Concentration (ng/mL)")
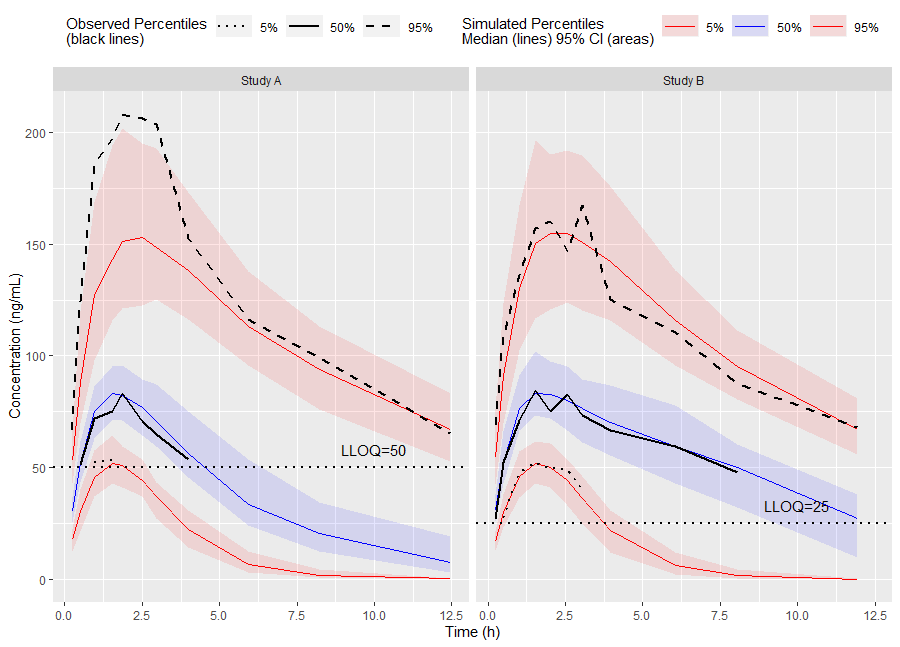
Or use the built-in plot() function from the tidyvpc package.
# Binless method using 10%, 50%, 90% quantiles and LOESS Prediction Corrected
# Add PRED variable to observed data from first replicate of sim_data
obs_data$PRED <- sim_data[REP == 1, PRED]
vpc <- observed(obs_data, x=TIME, y=DV) %>%
simulated(sim_data, y=DV) %>%
stratify(~ GENDER) %>%
predcorrect(pred=PRED) %>%
binless(loess.ypc = TRUE) %>%
vpcstats(qpred = c(0.1, 0.5, 0.9))
plot(vpc)