Description
Quantify the Contractile Nature of Vessels Monitored under an Operating Microscope.
Description
A variety of tools to allow the quantification of videos of the lymphatic vasculature taken under an operating microscope. Lymphatic vessels that have been injected with a variety of blue dyes can be tracked throughout the video to determine their width over time. Code is optimised for efficient processing of multiple large video files. Functions to calculate physiologically relevant parameters and generate graphs from these values are also included.
README.md
vmeasur
The goal of vmeasur is to quantify the contractile nature of vessels monitored under an operating microscope.
Installation
You can install the released version of vmeasur from CRAN with:
install.packages("vmeasur")
And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("JamesHucklesby/vmeasur")
Calibrating the operating microscope
To calibrate an operating microscope, take an image of a gradiated ruler. You can then use this function to calculate the number of pixels per mm.
calibrate_pixel_size()
Measuring the vessel diameter
Once video data is collected, the region of interest can be selected using select_roi. This provides a wizard that will assist the user through image analysis.
select_roi()
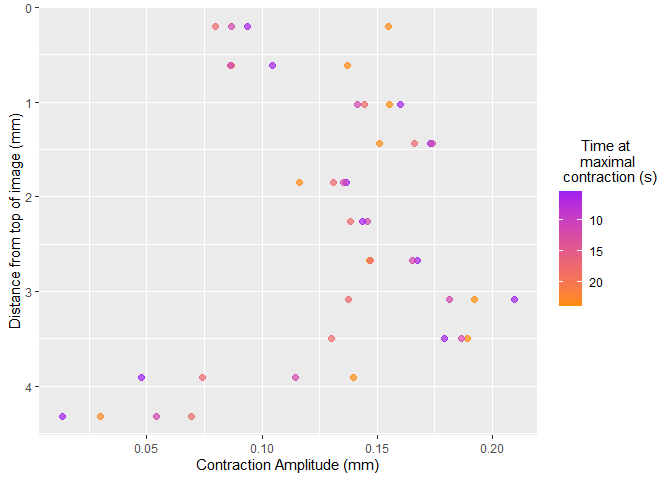
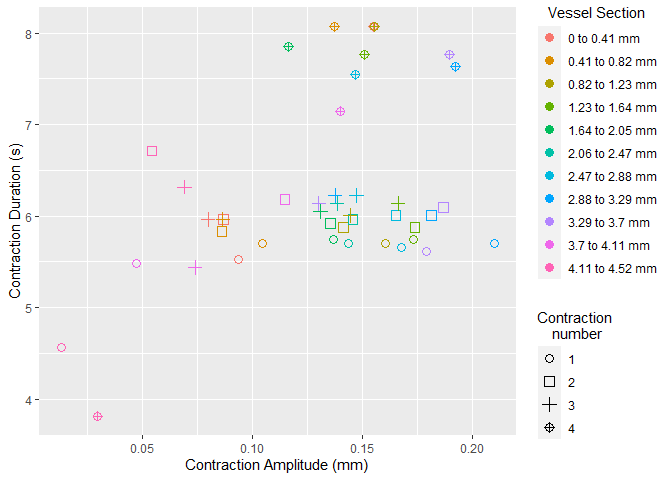
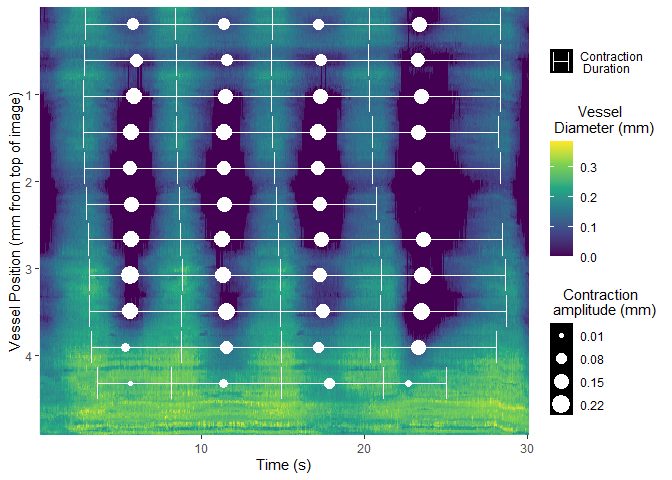
Once selected, vmeasur can output a variety of important parameters and graphs
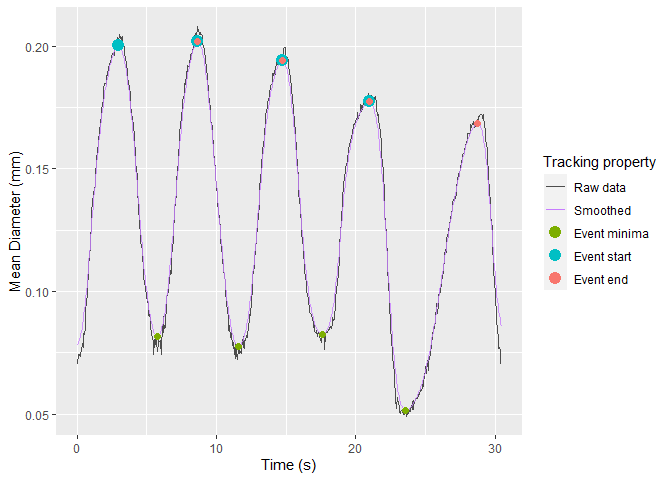
#> # A tibble: 4 x 12
#> # Rowwise:
#> event_maxima event_start event_end type start_value end_value max_value
#> <dbl> <dbl> <dbl> <chr> <dbl> <dbl> <dbl>
#> 1 531 472 647 contract 12.9 12.3 3.76
#> 2 261 195 331 contract 14.7 14.2 5.67
#> 3 130 67 195 contract 14.6 14.7 5.95
#> 4 396 331 472 contract 14.2 12.9 6.02
#> # ... with 5 more variables: baseline_change <dbl>, event_duration <dbl>,
#> # cont_duration <dbl>, fill_duration <dbl>, event_gradient <dbl>
#> # A tibble: 6 x 4
#> variable mean sd overall
#> <chr> <dbl> <dbl> <chr>
#> 1 CA 0.12 0.00645 0.12 (0.006448)
#> 2 CD 2.77 0.136 2.774 (0.1358)
#> 3 CS 0.0434 0.00390 0.04337 (0.003905)
#> 4 ED 6.36 0.908 6.36 (0.9081)
#> 5 EDD 0.193 0.0112 0.1932 (0.0112)
#> 6 EDD2 0.185 0.0152 0.1854 (0.01517)
#> X.1 y p_width excluded filename
#> 1 1 1 0 FALSE 1
#> 2 2 2 0 FALSE 1
#> 3 3 3 0 FALSE 1
#> 4 4 4 0 FALSE 1
#> 5 5 5 0 FALSE 1
#> 6 6 6 0 FALSE 1
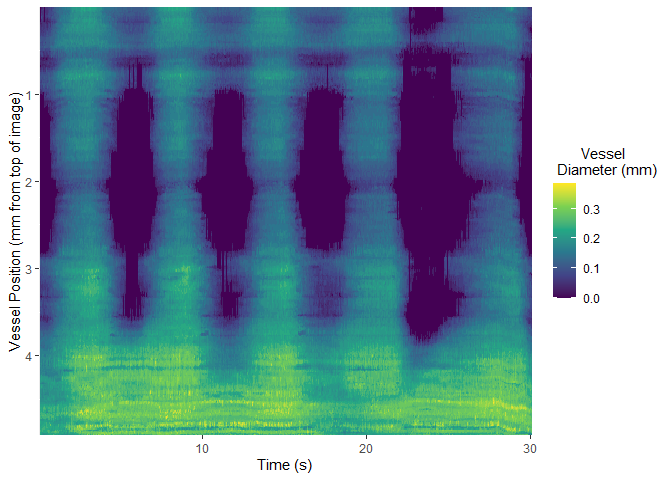
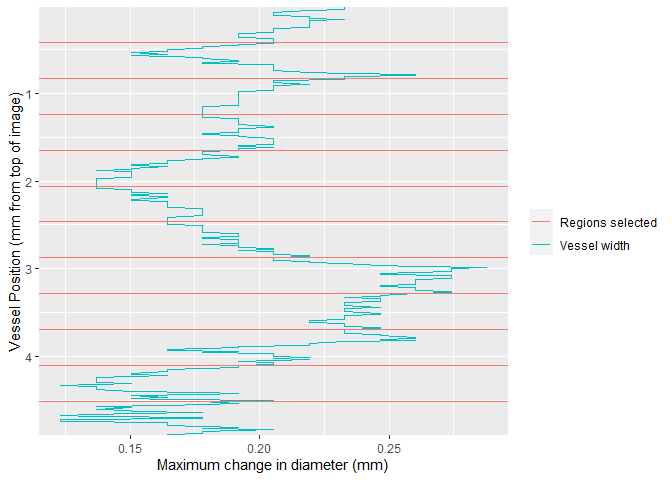
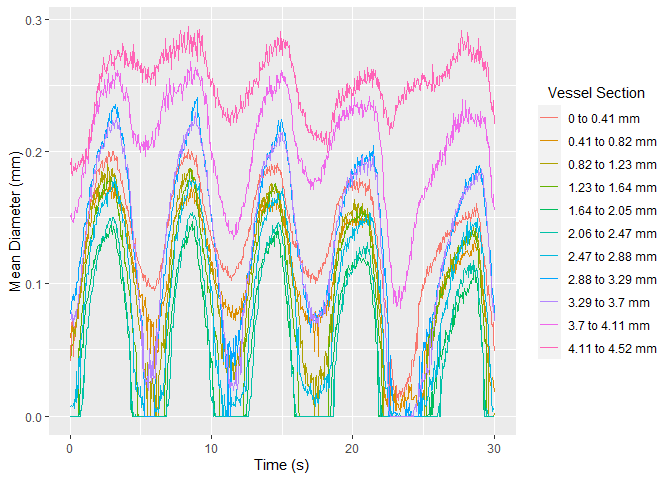
#> # A tibble: 6 x 15
#> # Groups: source_file [6]
#> ygroup event_maxima event_start event_end type start_value end_value
#> <chr> <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
#> 1 3 134 62 192 contract 13.2 13.2
#> 2 4 130 62 193 contract 12.6 12.7
#> 3 2 136 63 193 contract 12.3 12.1
#> 4 5 128 63 194 contract 9.97 9.88
#> 5 1 132 65 191 contract 14.2 14.2
#> 6 6 129 66 196 contract 10.5 10.6
#> # ... with 8 more variables: max_value <dbl>, baseline_change <dbl>,
#> # event_duration <dbl>, cont_duration <dbl>, fill_duration <dbl>,
#> # event_gradient <dbl>, source_file <dbl>, cont_id <int>
#> # A tibble: 6 x 5
#> ygroup variable mean sd overall
#> <chr> <chr> <dbl> <dbl> <chr>
#> 1 1 CA 0.104 0.0345 0.1038 (0.03449)
#> 2 1 CD 2.96 0.0913 2.961 (0.0913)
#> 3 1 CS 0.0349 0.0105 0.03486 (0.01051)
#> 4 1 ED 6.38 1.14 6.382 (1.145)
#> 5 1 EDD 0.188 0.00948 0.1875 (0.009478)
#> 6 1 EDD2 0.177 0.0190 0.1768 (0.01904)