Generate Random Data Sets.
wakefield
wakefield is designed to quickly generate random data sets. The user passes n (number of rows) and predefined vectors to the r_data_frame function to produce a dplyr::tbl_df object.

Table of Contents
Installation
To download the development version of wakefield:
Download the zip ball or tar ball, decompress and run R CMD INSTALL on it, or use the pacman package to install the development version:
if (!require("pacman")) install.packages("pacman")
pacman::p_load_gh("trinker/wakefield")
pacman::p_load(dplyr, tidyr, ggplot2)
Contact
You are welcome to: * submit suggestions and bug-reports at: https://github.com/trinker/wakefield/issues * send a pull request on: https://github.com/trinker/wakefield/ * compose a friendly e-mail to: [email protected]
Demonstration
Getting Started
The r_data_frame function (random data frame) takes n (the number of rows) and any number of variables (columns). These columns are typically produced from a wakefield variable function. Each of these variable functions has a pre-set behavior that produces a named vector of n length, allowing the user to lazily pass unnamed functions (optionally, without call parenthesis). The column name is hidden as a varname attribute. For example here we see the race variable function:
race(n=10)
## [1] Bi-Racial White Bi-Racial Native White White White Asian White Hispanic
## Levels: White Hispanic Black Asian Bi-Racial Native Other Hawaiian
attributes(race(n=10))
## $levels
## [1] "White" "Hispanic" "Black" "Asian" "Bi-Racial" "Native" "Other" "Hawaiian"
##
## $class
## [1] "variable" "factor"
##
## $varname
## [1] "Race"
When this variable is used inside of r_data_frame the varname is used as a column name. Additionally, the n argument is not set within variable functions but is set once in r_data_frame:
r_data_frame(
n = 500,
race
)
## Warning: `tbl_df()` is deprecated as of dplyr 1.0.0.
## Please use `tibble::as_tibble()` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_warnings()` to see where this warning was generated.
## # A tibble: 500 x 1
## Race
## <fct>
## 1 White
## 2 White
## 3 White
## 4 White
## 5 Black
## 6 Black
## 7 White
## 8 White
## 9 Hispanic
## 10 White
## # ... with 490 more rows
The power of r_data_frame is apparent when we use many modular variable functions:
r_data_frame(
n = 500,
id,
race,
age,
sex,
hour,
iq,
height,
died
)
## # A tibble: 500 x 8
## ID Race Age Sex Hour IQ Height Died
## <chr> <fct> <int> <fct> <times> <dbl> <dbl> <lgl>
## 1 001 White 25 Female 00:00:00 93 69 TRUE
## 2 002 White 80 Male 00:00:00 87 59 FALSE
## 3 003 White 60 Female 00:00:00 119 74 TRUE
## 4 004 Bi-Racial 54 Female 00:00:00 109 72 FALSE
## 5 005 White 75 Female 00:00:00 106 70 FALSE
## 6 006 White 54 Male 00:00:00 89 67 TRUE
## 7 007 Hispanic 67 Male 00:00:00 94 73 TRUE
## 8 008 Bi-Racial 86 Female 00:00:00 100 65 TRUE
## 9 009 Hispanic 56 Male 00:00:00 92 76 FALSE
## 10 010 Hispanic 52 Female 00:00:00 104 71 FALSE
## # ... with 490 more rows
There are 49 wakefield based variable functions to chose from, spanning R’s various data types (see ?variables for details).
| age | dice | hair | military | sex\_inclusive |
| animal | dna | height | month | smokes |
| answer | dob | income | name | speed |
| area | dummy | internet\_browser | normal | state |
| car | education | iq | political | string |
| children | employment | language | race | upper |
| coin | eye | level | religion | valid |
| color | grade | likert | sat | year |
| date\_stamp | grade\_level | lorem\_ipsum | sentence | zip\_code |
| death | group | marital | sex |
Available Variable Functions
However, the user may also pass their own vector producing functions or vectors to r_data_frame. Those with an n argument can be set by r_data_frame:
r_data_frame(
n = 500,
id,
Scoring = rnorm,
Smoker = valid,
race,
age,
sex,
hour,
iq,
height,
died
)
## # A tibble: 500 x 10
## ID Scoring Smoker Race Age Sex Hour IQ Height Died
## <chr> <dbl> <lgl> <fct> <int> <fct> <times> <dbl> <dbl> <lgl>
## 1 001 0.833 FALSE White 20 Female 00:00:00 92 69 TRUE
## 2 002 -0.529 TRUE Hispanic 83 Female 00:00:00 99 74 TRUE
## 3 003 -0.704 TRUE Hispanic 24 Male 00:00:00 115 62 TRUE
## 4 004 -0.839 TRUE Asian 19 Female 00:00:00 113 69 TRUE
## 5 005 0.606 TRUE White 70 Male 00:00:00 95 68 FALSE
## 6 006 1.46 FALSE Other 45 Female 00:00:00 110 78 FALSE
## 7 007 -0.681 TRUE Black 47 Female 00:00:00 98 64 TRUE
## 8 008 0.541 FALSE White 88 Male 00:30:00 75 70 TRUE
## 9 009 -0.294 FALSE Hispanic 89 Male 00:30:00 104 63 FALSE
## 10 010 0.0749 FALSE Hispanic 74 Female 00:30:00 105 69 TRUE
## # ... with 490 more rows
r_data_frame(
n = 500,
id,
age, age, age,
grade, grade, grade
)
## # A tibble: 500 x 7
## ID Age_1 Age_2 Age_3 Grade_1 Grade_2 Grade_3
## <chr> <int> <int> <int> <dbl> <dbl> <dbl>
## 1 001 67 24 89 82.4 86.8 90.6
## 2 002 55 76 27 87.3 85.4 89.8
## 3 003 60 61 22 82.2 87 90.1
## 4 004 50 19 56 96.4 86.6 95.6
## 5 005 83 77 71 88.8 87.5 84.4
## 6 006 55 71 76 87.3 96.5 86.5
## 7 007 88 36 75 92.1 91.6 93.4
## 8 008 71 48 81 87.9 91.4 80.9
## 9 009 76 78 21 86.9 93.6 84.3
## 10 010 49 68 47 85.5 93 86.6
## # ... with 490 more rows
While passing variable functions to r_data_frame without call parenthesis is handy, the user may wish to set arguments. This can be done through call parenthesis as we do with data.frame or dplyr::data_frame:
r_data_frame(
n = 500,
id,
Scoring = rnorm,
Smoker = valid,
`Reading(mins)` = rpois(lambda=20),
race,
age(x = 8:14),
sex,
hour,
iq,
height(mean=50, sd = 10),
died
)
## # A tibble: 500 x 11
## ID Scoring Smoker `Reading(mins)` Race Age Sex Hour IQ Height Died
## <chr> <dbl> <lgl> <int> <fct> <int> <fct> <times> <dbl> <dbl> <lgl>
## 1 001 2.48 FALSE 10 White 9 Male 00:00:00 93 44 TRUE
## 2 002 0.566 FALSE 14 Hispanic 10 Male 00:00:00 116 58 FALSE
## 3 003 -0.563 FALSE 19 Hispanic 8 Female 00:00:00 97 64 TRUE
## 4 004 0.0187 TRUE 19 White 9 Male 00:00:00 104 58 TRUE
## 5 005 -0.462 FALSE 17 Hispanic 11 Male 00:00:00 96 53 FALSE
## 6 006 -1.13 FALSE 17 White 10 Male 00:00:00 91 66 TRUE
## 7 007 -0.673 TRUE 15 White 13 Female 00:00:00 99 61 FALSE
## 8 008 0.164 TRUE 22 White 11 Male 00:00:00 106 47 FALSE
## 9 009 -0.227 FALSE 21 White 12 Female 00:00:00 101 54 TRUE
## 10 010 0.762 TRUE 22 White 8 Male 00:00:00 107 50 FALSE
## # ... with 490 more rows
Random Missing Observations
Often data contains missing values. wakefield allows the user to add a proportion of missing values per column/vector via the r_na (random NA). This works nicely within a dplyr/magrittr%>%then pipeline:
r_data_frame(
n = 30,
id,
race,
age,
sex,
hour,
iq,
height,
died,
Scoring = rnorm,
Smoker = valid
) %>%
r_na(prob=.4)
## # A tibble: 30 x 10
## ID Race Age Sex Hour IQ Height Died Scoring Smoker
## <chr> <fct> <int> <fct> <times> <dbl> <dbl> <lgl> <dbl> <lgl>
## 1 01 Hispanic 24 Female 01:30:00 92 70 NA NA NA
## 2 02 White NA Female <NA> NA NA FALSE 0.696 TRUE
## 3 03 Hispanic NA Female 02:00:00 107 68 FALSE -0.113 TRUE
## 4 04 Black 29 Female <NA> 93 75 TRUE -1.64 TRUE
## 5 05 <NA> 43 Female 03:30:00 NA NA NA -0.705 FALSE
## 6 06 Black NA <NA> 04:00:00 93 NA TRUE NA NA
## 7 07 Hispanic 60 <NA> <NA> NA NA TRUE NA NA
## 8 08 Hispanic NA <NA> <NA> NA NA TRUE NA FALSE
## 9 09 <NA> 34 <NA> 05:30:00 NA 70 NA -1.44 TRUE
## 10 10 White 88 <NA> <NA> NA NA NA NA NA
## # ... with 20 more rows
Repeated Measures & Time Series
The r_series function allows the user to pass a single wakefield function and dictate how many columns (j) to produce.
set.seed(10)
r_series(likert, j = 3, n=10)
## # A tibble: 10 x 3
## Likert_1 Likert_2 Likert_3
## * <ord> <ord> <ord>
## 1 Neutral Agree Agree
## 2 Strongly Agree Strongly Disagree Strongly Agree
## 3 Agree Strongly Disagree Agree
## 4 Disagree Strongly Disagree Agree
## 5 Neutral Strongly Agree Strongly Agree
## 6 Agree Disagree Disagree
## 7 Agree Agree Strongly Disagree
## 8 Agree Strongly Disagree Agree
## 9 Strongly Disagree Agree Neutral
## 10 Neutral Strongly Disagree Neutral
Often the user wants a numeric score for Likert type columns and similar variables. For series with multiple factors the as_integer converts all columns to integer values. Additionally, we may want to specify column name prefixes. This can be accomplished via the variable function’s name argument. Both of these features are demonstrated here.
set.seed(10)
as_integer(r_series(likert, j = 5, n=10, name = "Item"))
## # A tibble: 10 x 5
## Item_1 Item_2 Item_3 Item_4 Item_5
## <int> <int> <int> <int> <int>
## 1 3 4 4 4 5
## 2 5 1 5 3 1
## 3 4 1 4 5 4
## 4 2 1 4 4 5
## 5 3 5 5 2 5
## 6 4 2 2 3 4
## 7 4 4 1 4 1
## 8 4 1 4 1 2
## 9 1 4 3 5 3
## 10 3 1 3 5 5
r_series can be used within a r_data_frame as well.
set.seed(10)
r_data_frame(n=100,
id,
age,
sex,
r_series(likert, 3, name = "Question")
)
## # A tibble: 100 x 6
## ID Age Sex Question_1 Question_2 Question_3
## <chr> <int> <fct> <ord> <ord> <ord>
## 1 001 26 Male Strongly Agree Disagree Disagree
## 2 002 72 Male Disagree Agree Strongly Disagree
## 3 003 89 Male Strongly Disagree Strongly Disagree Strongly Agree
## 4 004 71 Female Agree Strongly Agree Disagree
## 5 005 56 Female Strongly Disagree Disagree Neutral
## 6 006 32 Female Strongly Disagree Strongly Agree Disagree
## 7 007 32 Female Strongly Disagree Strongly Agree Strongly Disagree
## 8 008 59 Female Neutral Strongly Agree Strongly Disagree
## 9 009 88 Male Agree Agree Agree
## 10 010 51 Male Agree Disagree Neutral
## # ... with 90 more rows
set.seed(10)
r_data_frame(n=100,
id,
age,
sex,
r_series(likert, 5, name = "Item", integer = TRUE)
)
## # A tibble: 100 x 8
## ID Age Sex Item_1 Item_2 Item_3 Item_4 Item_5
## <chr> <int> <fct> <int> <int> <int> <int> <int>
## 1 001 26 Male 5 2 2 4 5
## 2 002 72 Male 2 4 1 4 3
## 3 003 89 Male 1 1 5 4 4
## 4 004 71 Female 4 5 2 1 2
## 5 005 56 Female 1 2 3 3 2
## 6 006 32 Female 1 5 2 5 1
## 7 007 32 Female 1 5 1 1 5
## 8 008 59 Female 3 5 1 4 1
## 9 009 88 Male 4 4 4 3 2
## 10 010 51 Male 4 2 3 1 3
## # ... with 90 more rows
Related Series
The user can also create related series via the relate argument in r_series. It allows the user to specify the relationship between columns. relate may be a named list of or a short hand string of the form of "fM_sd" where:
fis one of (+, -, *, /)Mis a mean valuesdis a standard deviation of the mean value
For example you may use relate = "*4_1". If relate = NULL no relationship is generated between columns. I will use the short hand string form here.
Some Examples With Variation
r_series(grade, j = 5, n = 100, relate = "+1_6")
## # A tibble: 100 x 5
## Grade_1 Grade_2 Grade_3 Grade_4 Grade_5
## * <variable> <variable> <variable> <variable> <variable>
## 1 90.0 98.7 98.6 104.6 114.1
## 2 96.3 97.9 98.4 102.7 103.9
## 3 96.6 92.6 94.9 92.7 98.8
## 4 84.5 89.5 81.9 87.4 83.4
## 5 86.8 84.1 82.2 82.8 94.0
## 6 82.1 77.9 74.3 76.4 73.0
## 7 90.9 96.1 107.5 120.2 126.8
## 8 86.7 88.6 90.3 89.0 83.8
## 9 86.1 84.1 88.9 90.1 72.6
## 10 86.4 92.3 88.5 94.6 99.0
## # ... with 90 more rows
r_series(age, 5, 100, relate = "+5_0")
## # A tibble: 100 x 5
## Age_1 Age_2 Age_3 Age_4 Age_5
## * <variable> <variable> <variable> <variable> <variable>
## 1 83 88 93 98 103
## 2 48 53 58 63 68
## 3 80 85 90 95 100
## 4 46 51 56 61 66
## 5 33 38 43 48 53
## 6 53 58 63 68 73
## 7 34 39 44 49 54
## 8 31 36 41 46 51
## 9 81 86 91 96 101
## 10 50 55 60 65 70
## # ... with 90 more rows
r_series(likert, 5, 100, name ="Item", relate = "-.5_.1")
## # A tibble: 100 x 5
## Item_1 Item_2 Item_3 Item_4 Item_5
## * <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 0 -1 -1 -2
## 2 3 3 2 2 2
## 3 4 3 3 3 3
## 4 3 2 1 0 0
## 5 3 3 3 3 3
## 6 5 4 3 2 1
## 7 4 3 2 1 1
## 8 1 0 -1 -2 -2
## 9 3 2 1 1 1
## 10 1 0 0 -1 -2
## # ... with 90 more rows
r_series(grade, j = 5, n = 100, relate = "*1.05_.1")
## # A tibble: 100 x 5
## Grade_1 Grade_2 Grade_3 Grade_4 Grade_5
## * <variable> <variable> <variable> <variable> <variable>
## 1 90.8 90.80 99.880 109.8680 109.8680
## 2 89.8 80.82 80.820 64.6560 58.1904
## 3 90.3 99.33 109.263 109.2630 98.3367
## 4 95.2 76.16 91.392 91.3920 100.5312
## 5 89.1 98.01 117.612 105.8508 105.8508
## 6 86.8 95.48 95.480 114.5760 160.4064
## 7 93.4 93.40 93.400 102.7400 123.2880
## 8 92.7 83.43 91.773 110.1276 121.1404
## 9 84.9 93.39 93.390 102.7290 113.0019
## 10 84.7 84.70 93.170 93.1700 111.8040
## # ... with 90 more rows
Adjust Correlations
Use the sd command to adjust correlations.
round(cor(r_series(grade, 8, 10, relate = "+1_2")), 2)
## Grade_1 Grade_2 Grade_3 Grade_4 Grade_5 Grade_6 Grade_7 Grade_8
## Grade_1 1.00 0.84 0.57 0.41 0.31 0.30 0.16 0.15
## Grade_2 0.84 1.00 0.86 0.73 0.71 0.70 0.52 0.50
## Grade_3 0.57 0.86 1.00 0.93 0.92 0.90 0.77 0.71
## Grade_4 0.41 0.73 0.93 1.00 0.93 0.89 0.76 0.66
## Grade_5 0.31 0.71 0.92 0.93 1.00 0.93 0.83 0.79
## Grade_6 0.30 0.70 0.90 0.89 0.93 1.00 0.93 0.92
## Grade_7 0.16 0.52 0.77 0.76 0.83 0.93 1.00 0.95
## Grade_8 0.15 0.50 0.71 0.66 0.79 0.92 0.95 1.00
round(cor(r_series(grade, 8, 10, relate = "+1_0")), 2)
## Grade_1 Grade_2 Grade_3 Grade_4 Grade_5 Grade_6 Grade_7 Grade_8
## Grade_1 1 1 1 1 1 1 1 1
## Grade_2 1 1 1 1 1 1 1 1
## Grade_3 1 1 1 1 1 1 1 1
## Grade_4 1 1 1 1 1 1 1 1
## Grade_5 1 1 1 1 1 1 1 1
## Grade_6 1 1 1 1 1 1 1 1
## Grade_7 1 1 1 1 1 1 1 1
## Grade_8 1 1 1 1 1 1 1 1
round(cor(r_series(grade, 8, 10, relate = "+1_20")), 2)
## Grade_1 Grade_2 Grade_3 Grade_4 Grade_5 Grade_6 Grade_7 Grade_8
## Grade_1 1.00 -0.11 0.14 -0.21 -0.42 -0.29 -0.30 -0.27
## Grade_2 -0.11 1.00 0.49 0.44 0.18 0.24 0.23 0.51
## Grade_3 0.14 0.49 1.00 0.86 0.48 0.59 0.70 0.81
## Grade_4 -0.21 0.44 0.86 1.00 0.63 0.76 0.76 0.87
## Grade_5 -0.42 0.18 0.48 0.63 1.00 0.92 0.85 0.79
## Grade_6 -0.29 0.24 0.59 0.76 0.92 1.00 0.91 0.89
## Grade_7 -0.30 0.23 0.70 0.76 0.85 0.91 1.00 0.93
## Grade_8 -0.27 0.51 0.81 0.87 0.79 0.89 0.93 1.00
round(cor(r_series(grade, 8, 10, relate = "+15_20")), 2)
## Grade_1 Grade_2 Grade_3 Grade_4 Grade_5 Grade_6 Grade_7 Grade_8
## Grade_1 1.00 0.48 0.47 0.63 0.58 0.66 0.35 0.18
## Grade_2 0.48 1.00 0.90 0.87 0.54 0.43 0.67 0.23
## Grade_3 0.47 0.90 1.00 0.81 0.63 0.53 0.74 0.30
## Grade_4 0.63 0.87 0.81 1.00 0.75 0.72 0.71 0.47
## Grade_5 0.58 0.54 0.63 0.75 1.00 0.88 0.57 0.42
## Grade_6 0.66 0.43 0.53 0.72 0.88 1.00 0.68 0.54
## Grade_7 0.35 0.67 0.74 0.71 0.57 0.68 1.00 0.77
## Grade_8 0.18 0.23 0.30 0.47 0.42 0.54 0.77 1.00
Visualize the Relationship
dat <- r_data_frame(12,
name,
r_series(grade, 100, relate = "+1_6")
)
dat %>%
gather(Time, Grade, -c(Name)) %>%
mutate(Time = as.numeric(gsub("\\D", "", Time))) %>%
ggplot(aes(x = Time, y = Grade, color = Name, group = Name)) +
geom_line(size=.8) +
theme_bw()

Expanded Dummy Coding
The user may wish to expand a factor into j dummy coded columns. The r_dummy function expands a factor into j columns and works similar to the r_series function. The user may wish to use the original factor name as the prefix to the j columns. Setting prefix = TRUE within r_dummy accomplishes this.
set.seed(10)
r_data_frame(n=100,
id,
age,
r_dummy(sex, prefix = TRUE),
r_dummy(political)
)
## # A tibble: 100 x 8
## ID Age Sex_Male Sex_Female Democrat Republican Libertarian Green
## <chr> <int> <int> <int> <int> <int> <int> <int>
## 1 001 26 1 0 0 0 1 0
## 2 002 72 1 0 1 0 0 0
## 3 003 89 1 0 0 1 0 0
## 4 004 71 0 1 1 0 0 0
## 5 005 56 0 1 0 1 0 0
## 6 006 32 0 1 0 1 0 0
## 7 007 32 0 1 1 0 0 0
## 8 008 59 0 1 0 1 0 0
## 9 009 88 1 0 0 1 0 0
## 10 010 51 1 0 0 1 0 0
## # ... with 90 more rows
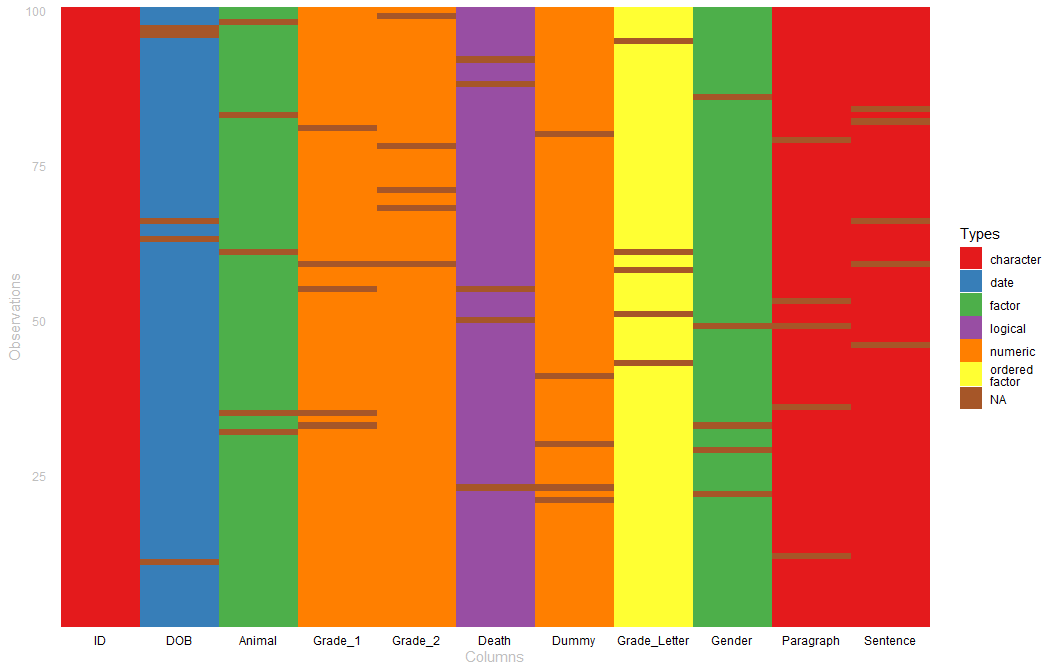
Visualizing Column Types
It is helpful to see the column types and NAs as a visualization. The table_heat (also the plot method assigned to tbl_df as well) can provide visual glimpse of data types and missing cells.
set.seed(10)
r_data_frame(n=100,
id,
dob,
animal,
grade, grade,
death,
dummy,
grade_letter,
gender,
paragraph,
sentence
) %>%
r_na() %>%
plot(palette = "Set1")