Statistical Methods for Life Data Analysis.
weibulltools 
Overview
The {weibulltools} package focuses on statistical methods and visualizations that are often used in reliability engineering. It provides a compact and easily accessible set of methods and visualization tools that make the examination and adjustment as well as the analysis and interpretation of field data (and bench tests) as simple as possible.
Besides the well-known Weibull analysis, the package supports multiple lifetime distributions and also contains Monte Carlo methods for the correction and completion of imprecisely recorded or unknown lifetime characteristics.
Plots are created statically {ggplot2} or interactively {plotly} and can be customized with functions of the respective visualization package.
Installation
The latest released version of {weibulltools} from CRAN can be installed with:
install.packages("weibulltools")
Development version
Install the development version of {weibulltools} from GitHub to use new features or to get a bug fix.
# install.packages("devtools")
devtools::install_github("Tim-TU/weibulltools")
Usage
Getting started
Create consistent reliability data with columns:
x- lifetime characteristicstatus- binary data (0 for censored units and 1 for failed units)id(optional) - identifier for units
library(weibulltools)
rel_tbl <- reliability_data(data = shock, x = distance, status = status)
rel_tbl
#> Reliability Data with characteristic x: 'distance':
#> # A tibble: 38 × 3
#> x status id
#> <int> <dbl> <chr>
#> 1 6700 1 ID1
#> 2 6950 0 ID2
#> 3 7820 0 ID3
#> 4 8790 0 ID4
#> 5 9120 1 ID5
#> # … with 33 more rows
Probability estimation and visualization
Estimation of failure probabilities using different non-parametric methods:
prob_tbl <- estimate_cdf(x = rel_tbl, methods = c("mr", "kaplan", "johnson", "nelson"))
#> The 'mr' method only considers failed units (status == 1) and does not retain intact units (status == 0).
prob_tbl
#> CDF estimation for methods 'mr', 'kaplan', 'johnson', 'nelson':
#> # A tibble: 125 × 6
#> id x status rank prob cdf_estimation_method
#> <chr> <int> <dbl> <dbl> <dbl> <chr>
#> 1 ID1 6700 1 1 0.0614 mr
#> 2 ID5 9120 1 2 0.149 mr
#> 3 ID13 12200 1 3 0.237 mr
#> 4 ID15 13150 1 4 0.325 mr
#> 5 ID19 14300 1 5 0.412 mr
#> # … with 120 more rows
Visualization of the obtained results in a distribution-specific probability plot:
prob_vis <- plot_prob(x = prob_tbl, distribution = "weibull",
title_main = "Weibull Probability Plot",
title_x = "Distance",
title_y = "Probability of Failure in %",
title_trace = "Methods",
plot_method = "gg")
prob_vis
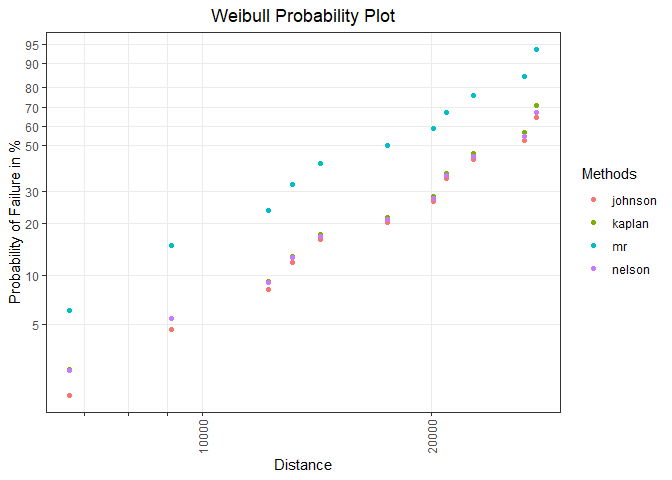
Model estimation and visualization
Parametric model estimation with respect to the used methods:
rr_list <- rank_regression(x = prob_tbl, distribution = "weibull")
rr_list
#> List of 4 model estimations:
#> Rank Regression
#> Coefficients:
#> mu sigma
#> 10.2596 0.3632
#> Method of CDF Estimation: johnson
#>
#> Rank Regression
#> Coefficients:
#> mu sigma
#> 10.2333 0.3773
#> Method of CDF Estimation: kaplan
#>
#> Rank Regression
#> Coefficients:
#> mu sigma
#> 9.8859 0.3956
#> Method of CDF Estimation: mr
#>
#> Rank Regression
#> Coefficients:
#> mu sigma
#> 10.2585 0.3852
#> Method of CDF Estimation: nelson
Model visualization in an existing probability plot:
mod_vis <- plot_mod(p_obj = prob_vis, x = rr_list, distribution = "weibull",
title_trace = "RR")
mod_vis
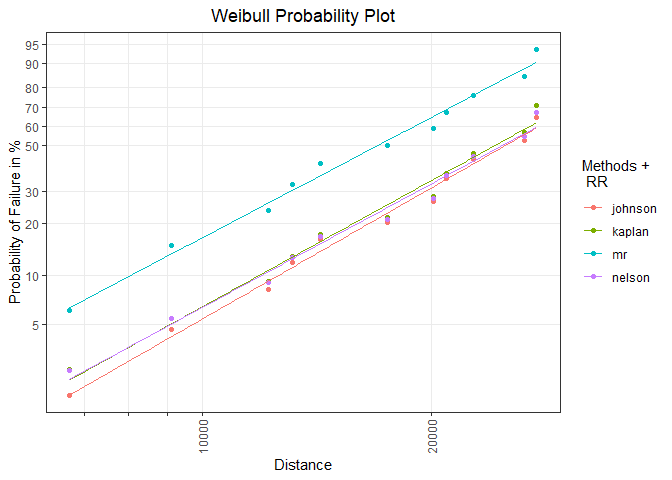
Getting help
If you notice a bug or have suggestions for improvements, please submit an issue with a minimal reproducible example on GitHub. For further questions, please contact Tim-Gunnar Hensel.
